PCA and Whitening on natural images
In this exercise, you will implement PCA, PCA whitening and ZCA whitening, and apply them to image patches taken from natural images.
You will build on the MATLAB starter code which we have provided in the Github repository You need only write code at the places indicated by YOUR CODE HERE in the files. The only file you need to modify is pca_gen.m.
Step 0: Prepare data
Step 0a: Load data
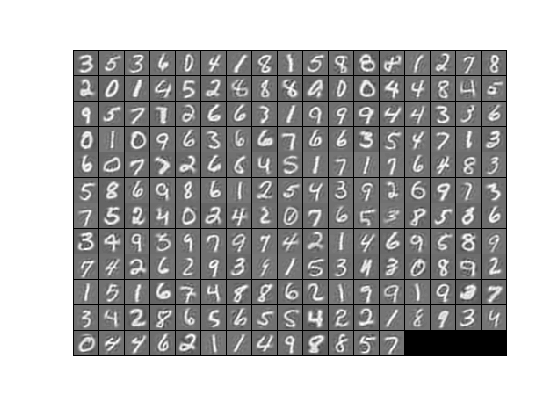
The starter code contains code to load a set of MNIST images. The raw patches will look something like this:

These patches are stored as column vectors
Step 0b: Zero mean the data
First, for each image patch, compute the mean pixel value and subtract it from that image, this centering the image around zero. You should compute a different mean value for each image patch.
Step 1: Implement PCA
Step 1a: Implement PCA
In this step, you will implement PCA to obtain
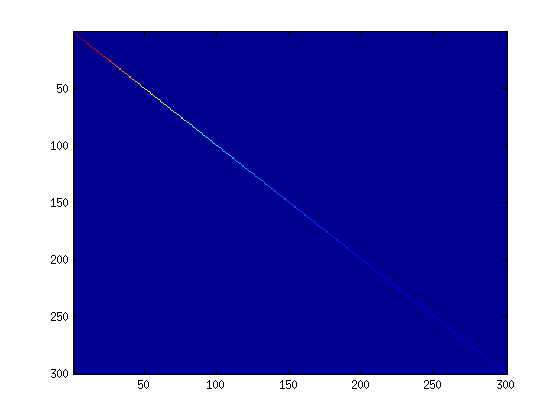
Step 1b: Check covariance
To verify that your implementation of PCA is correct, you should check the covariance matrix for the rotated data imagesc. The image should show a coloured diagonal line against a blue background. For this dataset, because of the range of the diagonal entries, the diagonal line may not be apparent, so you might get a figure like the one show below, but this trick of visualizing using imagesc will come in handy later in this exercise.
Step 2: Find number of components to retain
Next, choose
Step 3: PCA with dimension reduction
Now that you have found
To see the effect of dimension reduction, go back from
 |
 |
|
| Raw images |
PCA dimension-reduced images (99% variance) |
PCA dimension-reduced images (90% variance) |
Step 4: PCA with whitening and regularization
Step 4a: Implement PCA with whitening and regularization
Now implement PCA with whitening and regularization to produce the matrix
epsilon = 0.1
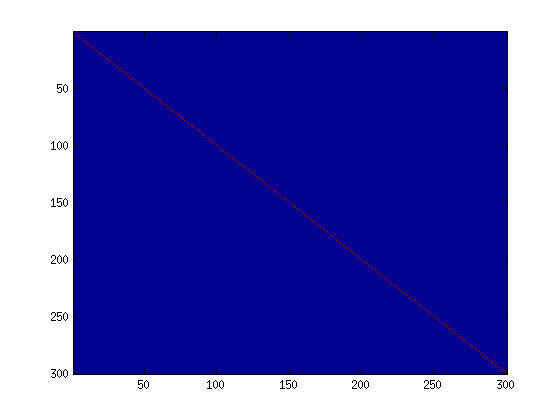
Step 4b: Check covariance
Similar to using PCA alone, PCA with whitening also results in processed data that has a diagonal covariance matrix. However, unlike PCA alone, whitening additionally ensures that the diagonal entries are equal to 1, i.e. that the covariance matrix is the identity matrix.
That would be the case if you were doing whitening alone with no regularization. However, in this case you are whitening with regularization, to avoid numerical/etc. problems associated with small eigenvalues. As a result of this, some of the diagonal entries of the covariance of your
To verify that your implementation of PCA whitening with and without regularization is correct, you can check these properties. Implement code to compute the covariance matrix and verify this property. (To check the result of PCA without whitening, simply set epsilon to 0, or close to 0, say 1e-10). As earlier, you can visualise the covariance matrix with imagesc. When visualised as an image, for PCA whitening without regularization you should see a red line across the diagonal (corresponding to the one entries) against a blue background (corresponding to the zero entries); for PCA whitening with regularization you should see a red line that slowly turns blue across the diagonal (corresponding to the 1 entries slowly becoming smaller).
|
|
Step 5: ZCA whitening
Now implement ZCA whitening to produce the matrix epsilon set to 1, 0.1, and 0.01, and see what you obtain. The example shown below (left image) was obtained with epsilon = 0.1.
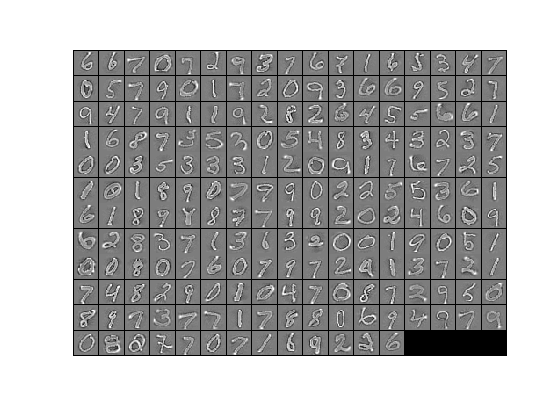
|

|
| ZCA whitened images | Raw images |